eopegwas
Maternal Cardiovascular-Related Single Nucleotide Polymorphisms, Genes and Pathways Associated with Early-Onset Preeclampsia
Maternal Cardiovascular-Related Single Nucleotide Polymorphisms, Genes and Pathways Associated with Early-Onset Preeclampsia
This repository holds the code for the paper.
You can find more information and data on the OSF project page: https://osf.io/ecpw6/
Results
The paper (todo: add link to paper) contains a summary and explanation of the findings.
The src folder contains the scripts used to do the analysis.
You can also skim the R exploratory analysis of the data, without running it yourself, in notebook format here: https://breckuh.github.io/eopegwas/src/main.nb.html
Results of snpQC report are here: https://github.com/breckuh/preeclampsiagwas/blob/master/snpQCreport.pdf
How to Use This Code
If you want, you can reuse this code with your own data to perform similar experiments.
You can also use it with the provided simulated data (in mockData/) which is identical to our data except simulated for privacy reasons.
If you’d like to run this code and get the exact same results as us, please contact us for our raw data files.
The data sources required are:
- data/072312_FinalReport.txt
- data/clinical.csv
- data/cardio-metabo_chip_11395247_a.csv
The format for the clinical data is contained in eopClinicalSsv.grammar.
Setup Instructions
To run the R code in this repo:
git clone https://github.com/breckuh/eopegwas
cd eopegwas
mkdir ignore
mkdir cache
mkdir output
rmarkdown::render('main.rmd')
Pvals
For some of the R notebooks, you’ll first have to run thread.r to generate the pvals for each model you want to test. thread.r will save
results to the cache folder.
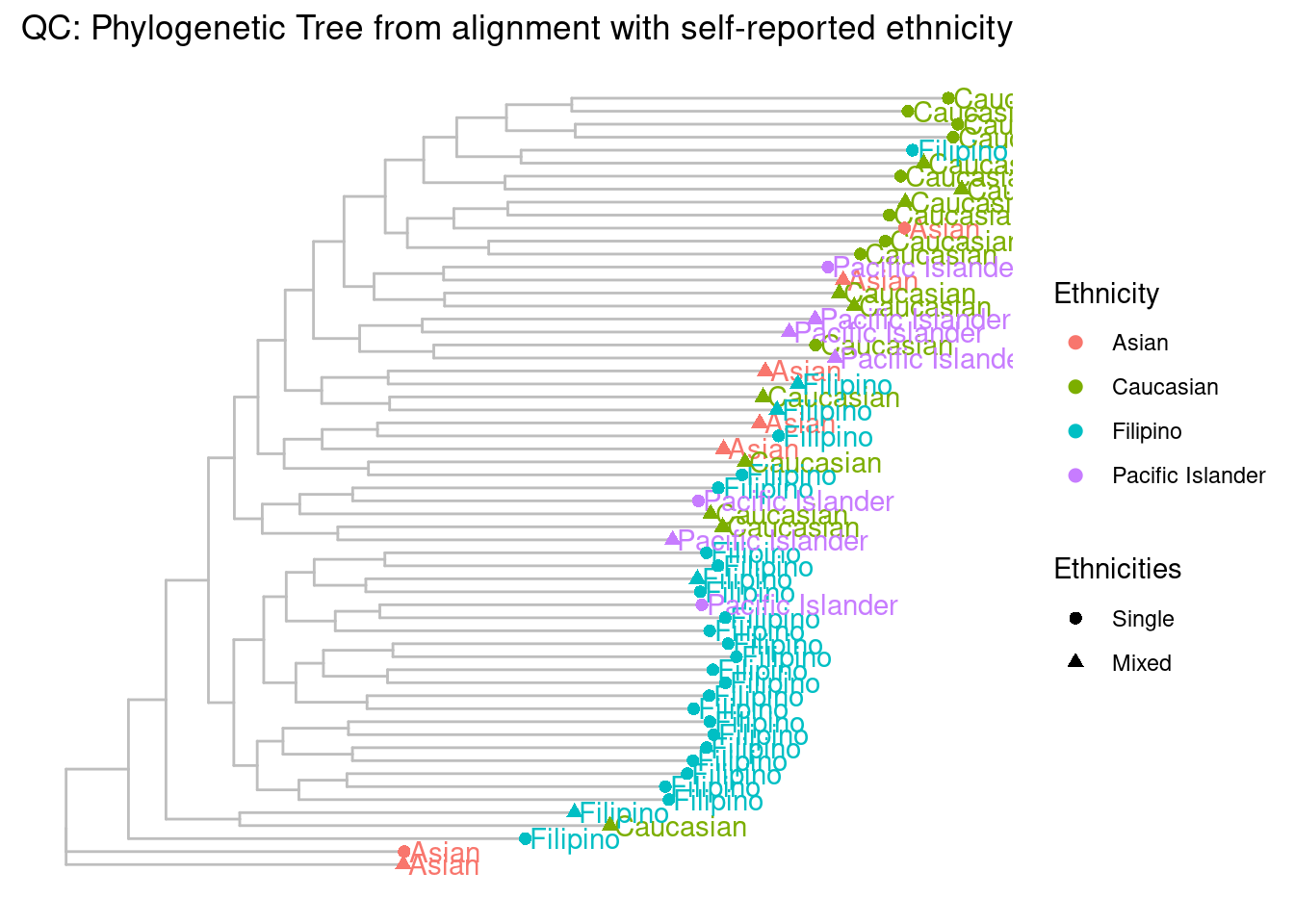
Phylotree
You’ll need to download and install this program: http://chibba.pgml.uga.edu/snphylo/ to run the phylotree.rmd script. Put the Phylotree scripts in the ignore folder.
Testing
Run rmarkdown::render('main.rmd') to execute the majority of the code